Maria Rubel's group studies the behavior of DNA nanosensors in detecting complex fragments: fragments with stable secondary stacking, repetitive sites, highly variable or guanine cytosine-rich sites. Maria's group is also collecting and systematizing previously accumulated material, forming a database to train artificial intelligence that will design DNA nanosensors in future. This work provides a better understanding of how DNA nanosensors function and prepares them to move into diagnostic practice.
Muhannad Ateiah’s group aims to development highly sensitive DNA nanoconstructs for the amplification-free detection of nucleic acids. Being the most sensitive probe among protein enzyme-free approaches, BiDz probes are still not sensitive enough to detect the majority of biological DNA or RNA analytes without amplification. Our efforts to increase the Limit of detection (LOD) of BiDz probes led to a design would increase the diffusion rate of F-sub to the core of the activated BiDz probe, so called DNA nanomachine (Figure 1). We aim to further improve this design by increasing the number of cleaving elements, increasing the number of analyte-binding arms and further increasing of the diffusion rate of F-sub. As a result of the implementation of these modifications, we believe that the developed DNA nanoconstruct can increase the detection efficiency to attomolar concentrations, which corresponds to the concentration of RNA in cells. The developed DNA nanoconstruct can be used to create diagnostic devices that can be used for point-of-care diagnostics and in circumstances where amplification step is not possible. In addition, this approach can become the basis for the development of an automatic device for diagnostic of pathogens and other applied purposes.
Another field of research — development of fluorescent hybridization probes for highly selective detection of double stranded DNA analytes. Specific binding of double stranded (ds) DNA by hybridization probe under physiological conditions can be used for gene knockdown, chromosome mapping in living cells, detection of important diagnostic targets, and ultimately result in novel therapeutic intervention strategies against genetic diseases. We aim to develop such constructs using DNAzymes and Locked Nucleic Acids (LNA) modifications. All constructs go through two stages of development: optimizing the number and position of LNA modifications regarding to the catalytic core and testing the constructs under near physiological conditions and at different temperatures. By achieving our goal, we hope to advance the future of hybridization probes by its evolvement into DNA machines as a more sophisticated sensing and specialized tool.
Daria Gorbenko's group is engaged in the optimization of some types of DNA sensors, mainly 4-way-junction (4WJ) probes and multicomponent peroxidase machines. The main goal of the group is the integration of existing and in vitro proved to be effective sensors into compact detection systems. Key areas for today:
- adoption of chemiluminescent DNA sensors into a microfluidic sensor;
- immobilization and storage of DNA sensors on solid substrates (nitrocellulose membranes, modified glasses);
- bioconjugation of DNA sensors on nanoparticles for subsequent detection in systems for quantitative signal counting / visual aggregation of nanoparticles as a qualitative indicator of the detection of the target analyzed agent.
Tatyana Zemerova's group is developing a test system for genotyping strains of Mycobacterium tuberculosis. The main problem in the treatment of tuberculosis is the resistance to antibiotics of different bacterial strains, therefore, the development of a system that allows genotyping strains quickly and without large costs can significantly increase the effectiveness of treatment. The strains differ from each other in the presence/absence of mutations - single nucleotide polymorphisms (SNPs), and development is aimed at detecting the presence/absence of SNPs. The object of the study is the toxin-antitoxin system functioning in bacteria in order to avoid the action of antibiotics. The genotyping test system being developed consists of SNP-sensitive DNA machines, and our goal is the direct detection of a double-stranded target site. Also, our goal is to develop a multiplex system that allows detection of up to three samples in one reaction solution, which will further reduce the time and complexity of the analysis.
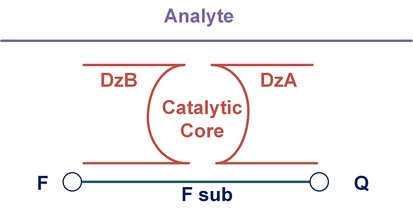
Fig. 1. Binary deoxyribozyme design.
External consultant - Dmitry Kolpashchikov.