Project 1: Elucidating the underpinnings of apomixis, a major force in evolution and implications for agriculture.
Sexuality produces a wide variety of new forms and features in living organisms. Often during hybridization heterosis (emergence of hybrid vigour) occurs in the first generation of hybrids. In plants, heterosis is accompanied by an increase in yield, improved nutritional properties of seeds and fruits, or qualitatively better agricultural plant characteristics. However, in subsequent generations, valuable phenotypic traits are lost following meiosis, recombination, and segregation, and the plants lose the advantages conferred by hybridization and heterosis. Apomixis is a process by which plants produce offspring that are genetically identical to the parent through asexually produced seeds, which could be found in more than 400 plant species representing almost 40 families. Apomixis could be considered as a developmental variation of sexual reproduction in which some steps are lost, reduced, deregulated or desynchronized (Fig.1). The main features of gametophytic apomixis are:
- Avoidance of meiosis (apomeiosis);
- Embryo formation via parthenogenesis;
- Functional endosperm develops either autonomously or pseudogamously (central cell fertilized by sperm cell).
The potential of apomixis as a next generation technology for plant breeding attracts huge interest of scientists to elucidate the molecular and genetic mechanisms of its regulation. Study of genetics and genomics of apomixis facilitates to understanding the role of heterozygosity in the transition from sexual to the asexual reproduction and also evolution of apomixis as a means of reproductive plasticity, which helps the survival of species.
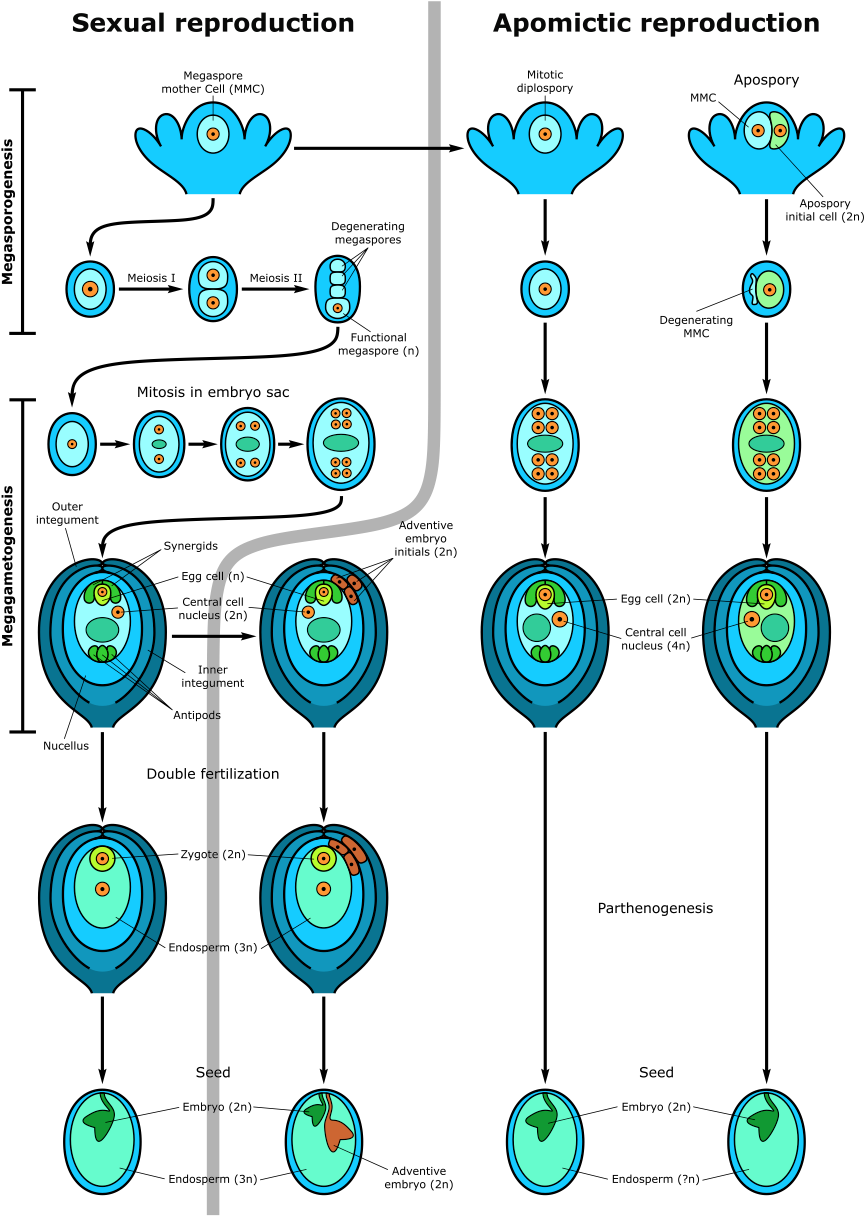
Fig. 1. Schematic representation of sexaual and apomictic development.
The left side of the figure shows the sexual pathway and sporophytic apomixis (adventive embryony). In the sexual pathway the megaspore mother cell (MMC) undergoes two meiotic divisions producing a tetrad of megaspores. Three megaspores degenerate while the functional megaspore gives rise to the embryo sac following three rounds of mitotic divisions (female gametophyte = megagametophyte). The mature embryo sac is 7-celled, 8-nucleate with the egg cell and synergid cells (egg cell apparatus) located at the micropylar pole of the embryo sac and three antipodal cells at the opposite pole. Two polar nuclei fuse to form the diploid nucleus of the central cell. After double fertilisation, one sperm fuses with the egg cell forming the diploid zygote, whereas the second sperm fertilises the central cell producing the first nuclei of triploid endosperm. Synergids participate in the perception of the sperm cells and burst after fertilisation. The mature seed consists of the diploid embryo, the triploid endosperm (nourishing tissue) and remnants of the maternal sporophytic nucellar and integumental tissues form the seed coat. An adventive embryo may develop from the nucellar or inner integumental sporophytic tissues and develop alongside the sexual embryo. (Adopted from Brukhin 2017). The Right part of the figure shows gametophytic apomixis. Meiosis is bypassed and consequently reduction is avoided. Embryo sac development is initiated from an unreduced diplospore (diplospory) or from an apospory initial cell (apospory). The embryo develops parthenogenetically from the unreduced egg cell while endosperm forms either autonomously or through fertilisation of the central cell (in case of pseudogamy). The mature apomictic seed contains the embryo with a relative ploidy level of 2n, while the endosperm could be of variable ploidy but usually not less than 4n.

Fig. 2 Boechera sp. plant and inflorescence
Closely related to the model plant Arabidopsis thaliana, the genus Boechera (Fig. 2) is known to contain both sexual and apomictic species or accessions. The genome of the latter are of hybridogenic origin and the structure of their chromosomes often affected by the consequences of hybridization, leading to alloploidy, aneuploidy, the replacement of homeologous chromosomes, and aberrant chromosomes. Unlike sexual counterparts, apomictic Boechera accessions are highly heterozygous (Fig. 3). In our research, we are attempting to de novo assemble and to annotate (identify the structural elements of the genome) the whole genomes and distinguish differences between sexual and apomictic accessions. We exploit the Illumina, Pacific Bioscience, Oxford Nanopore next-generation sequencing (NGS) platforms, perform fluorescence in situ hybridization (FISH) analysis in sexual and apomictic accessions. Apomixis usually occurs in polyploid perennial species, while diploid species of the same genus reproduce sexually. However, several diploid accessions of Boechera possess a high penetrance of apomixis. They have been the focus of genetic, cytological (Fig. 4), and ecological studies, due to its close relatedness to the model plant Arabidopsis thaliana, for which genetic and functional annotation data are very well established. We also perform phylogenetic and evolutionary analysis of the apomixis associated genes (Fig. 5). Assembly and correct annotation of highly heterozygous genomes of apomictic species of Boechera genomes will provide a basis to decipher the hybridogenetic events that led to the formation of apomictic accessions.
The study of genetics and genomics of apomixis facilitates to an understanding of the role of heterozygosity in the transition from sexual to the asexual reproduction and also the evolution of apomixis as a means of reproductive plasticity, which helps to the survival of the species.
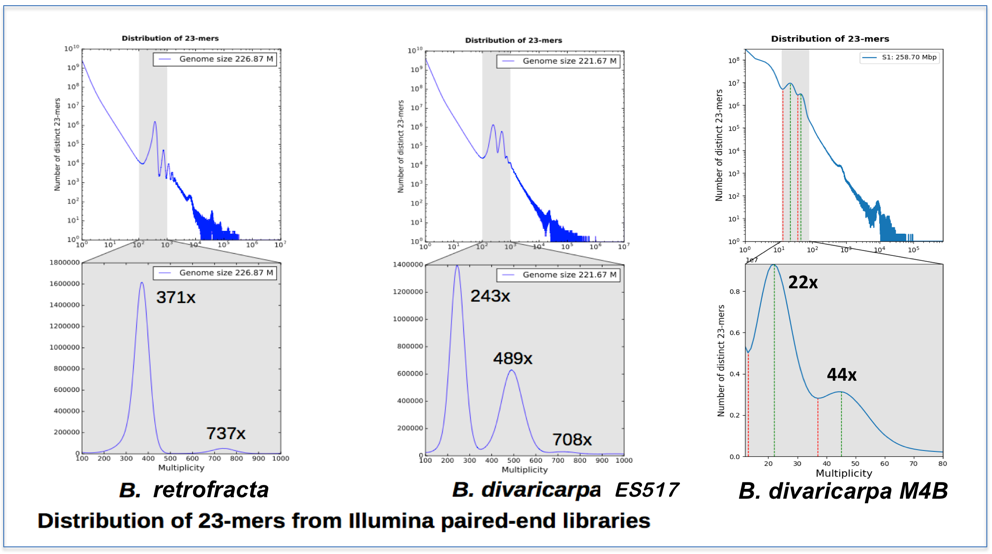
Fig. 3 Distribution of 23-mers from Illumina paired-end libraries. One distinct main peak in the spectrum of B. retrofracta (sexual accession) indicates a low level of heterozygosity in the species with sexual reproduction (left image). Comparable heights of the first and second peaks in two (right) apomictic accessions Boechera indicate a high level of heterozygosity in apomictic species.
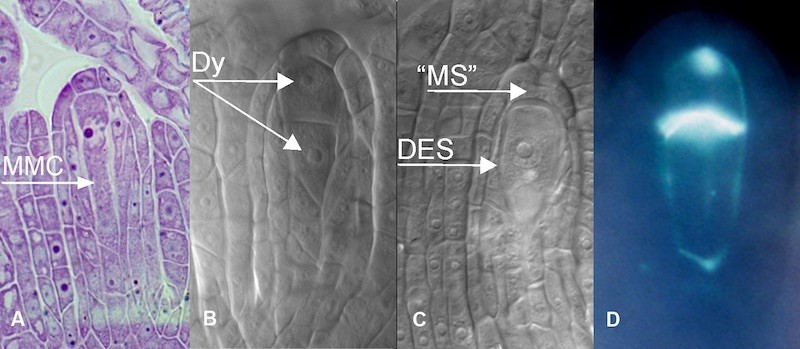
Fig. 4 Apomeiosis during megasporogenesis in B. holboellii s.l. A: Megaspore Mother Cell (MMC); B: Diplospory Diad (Dy); C: Single nuclear Diplosporic Embryo Sac (DES) with the remnants of "megaspore" (“MS”); D: callose in the cell wall of the diplosporic dyad
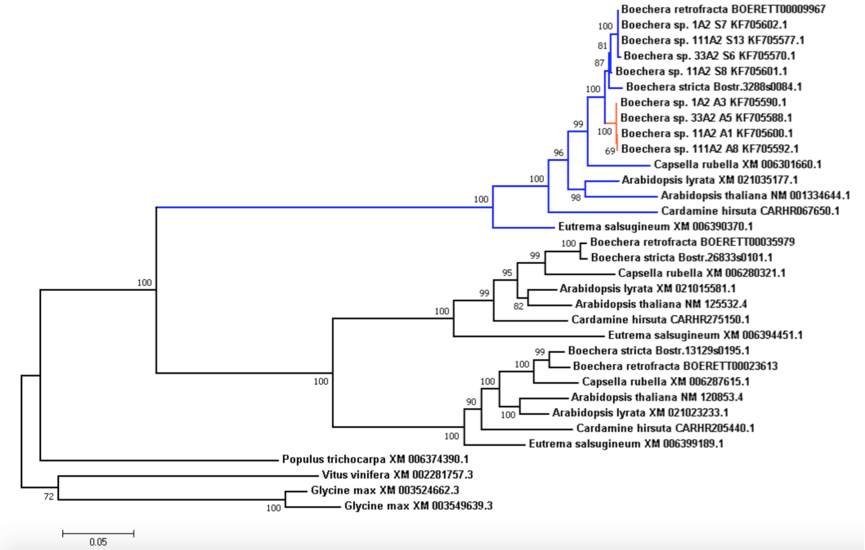
Fig. 5 Phylogenetic tree of the isoforms of APOLLO locus (exonuclease NEN) that is tightly associated with apomixis in Boechera. Seven Brassucaceae species were chosen to help to analyse evolution of homologs of APOLLO locus of apomictic Boechera species from Corral et al (2013). Sequences of Populus trichocarpa, Vitus vinifera and Glycine max were used as outgroup. The clade related to the APOLLO locus is shown in blue, with apo-alleles shown in red. Numbers near nodes represent corresponding bootstrap support. Branches in the tree are grouped by genes rather than by species, suggesting that the triplication event took place before the separation of the Brassicaceae species. Branch leading to apo-alleles is under positive selection (Ka/Ks 1.4646), which is typical for paralogues that are required to serve a novel function.
Understanding the genetic components that regulate apomixis is important for both plant breeding and evolution. The introduction of apomixis in cultivated plants could allow long-term fixation of complex genotypes, including F1 hybrids, which are often used in agriculture. The molecular and evolutionary mechanisms underlying apomixis are poorly understood. This project is aimed at studying the genetic mechanisms that control apomixis by comparative genomic study of sexually reproducing plants and closely related apomictic plants. Phylogenetic and evolutionary analyses are employed as a framework for comparisons.
In collaboration with St. Petersburg State University; University of Zurich, Switzerland; University of Brno, Czech Republic.
Project 2: Phylogenetic and functional analysis of genes associated with apomixis in representatives of the Boechera genus.
This project is aimed at studying the genetic mechanisms that control apomixis by identifying genomic and transcriptomic differences in sexual and apomictic plants, as well as functional analysis of candidate genes associated with apomixis and their phylogenetic and evolutionary analysis. An important and nontrivial task, which is still unexplored. During the project implementation it is expected to detect additional alleles of genes regulating meiosis and parthenogenesis; characterization of the structure of genes associated with apomixis, using data on gene copy numbers (CNVs) and polymorphisms (SNVs and InDels); determination of gene ontology classes enriched with apomictic alleles; obtaining data on the comparative expression of genes associated with apomixis; cloning the best candidate genes; and an analysis of complementation in Arabidopsis and, possibly, editing these genes in Boechera using CRISPR.
In collaboration with St. Petersburg State University; Komarov Botanical Institute, Russian Academy of Sciences; Çanakkale Onsekiz Mart University, Turkey.
Project 3: Development of a database of plant genomes and organisms
Create a searchable open access database of the plant whole genome assembled to chromosome scale or contig scale annotated genomes as well as yet unannotated genomes.