Daria Gorbenko and her colleagues from ITMO's SCAMT Institute. Photo by Dmitry Grigoryev / ITMO.NEWS
Nanorobot with arms
A DNA nanomachine is a small platform of about 20-30 nucleotides in size, equipped with special “arms” as the scientists call them. Approaching an RNA structure, the robot-like device tightly bind onto the elaborately folded nucleic acids with its arms so that the RNA chain starts to unfold. The procedure is made possible by the principle of complementarity. This property allows nucleic acids to attract each other and fuse with their complementary pairs (adenine-thymine and guanine-cytosine).
The proposed unfolding method eliminates the need for thermal amplification – a complex procedure that must be performed in specific laboratory conditions and using thermal cyclers. Using this technology, researchers can elicit this reaction (or hybridization, meaning the formation of double-stranded nucleic acids) at room temperature, which makes the new technique favorably different from standard PCR testing.
“RNA and DNA structures are typically folded and only change their state when exposed to high temperatures, which makes it difficult to detect target fragments. Our nanomachines can locate the desired areas – analytes of single-stranded RNA viruses – without using additional equipment and heating the nucleic acids . The process goes like this: the nanobots approach their target, wrap their arms around it, and make it unfold,” explains Daria Gorbenko, a staff member at ITMO's SCAMT Institute and the first author of the paper.
A custom platform is developed for each particular pathogen, designed specifically to prevent undesired crosstalk between the machine’s structure elements and RNA analytes. However, it is enough to create a platform once for each pathogen. The researchers already have Cytomegalovirus (a herpes virus), Haemophilus influenzae (a pneumonia pathogen), and Listeria monocytogenes (a food-borne pathogen) in their database.
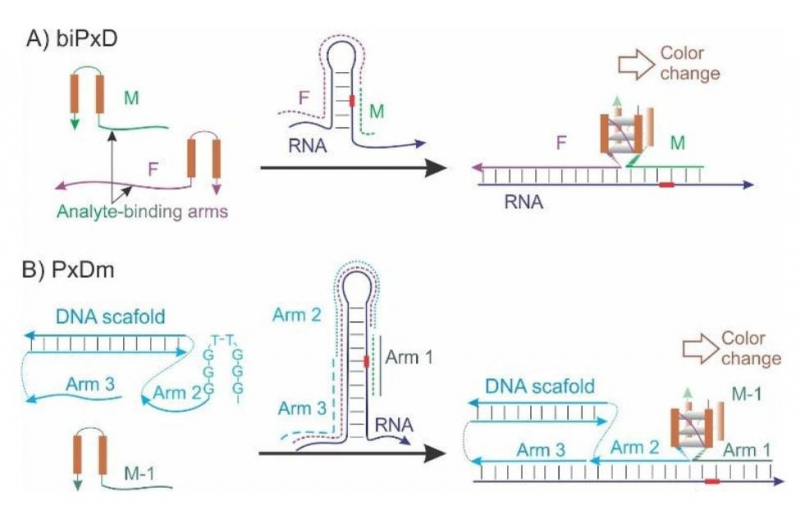
The difference between binary and multi-component machines proposed in the study. Photo courtesy of the authors
Unique design
As noted by the researchers, the main advantage of their research is not the technology alone (it is already widely used worldwide) but the design of their machines, too. The binding of the machine’s arms with an analyte (a pathogen-specific piece of RNA) leads to the formation of G-quadruplex structures (3D structures formed by four nucleic acid strands). When additional reagents are added, the normally transparent solution changes its color. The developers believe such a colorimetric reaction makes the new testing method better than PCR tests.
“The properties of G-quadruplexes offer a scope of opportunities for developing a simple and evident testing system that changes color upon reaction with reagents. For example, diaminobenzidine, hemin and hydrogen peroxide make it turn dark-brown. While PCR tests cannot show results without special equipment, our test visually demonstrates whether the detection has occurred or not,” says Daria Gorbenko.
Although the entire testing process, from taking material and extracting nucleic acid to the formation of a G-quadruplex and change in color, takes from two to three hours, the researchers are certain that the procedure can be accelerated. In the future, they plan to embed their device in a small sealed package to make testing possible even at home.

Photo by Dmitry Grigoryev / ITMO.NEWS
Accidental breakthrough
The study was published in Chemical Communications as one of the ten top-priority publications of the year. As the researchers admit, the major discovery that lies at the heart of this research was made by accident and is yet to be fully studied. During their research, the scientists noticed that their method is applicable not only to single-stranded RNA but also to firmly hybridized double-stranded DNA analytes, which has never been unfolded before without annealing.
“We’ve discovered something that we cannot still fully understand. We created and used our nanomachine for single-stranded RNA, that is, those molecules that can easily bind with the machine’s single-stranded arms due to their complementarity. These structures indeed have a favorable thermodynamic interaction potential. And the fact that our probes can detect a double-stranded analyte without additional manipulations like annealing came as a surprise to us. As far as we know, this has never been done before. We cannot say for sure why it happens but we have many hypotheses, which we’re testing right now,” comments Daria Gorbenko.

Daria Gorbenko. Photo by Dmitry Grigoryev / ITMO.NEWS
Reference: Daria Gorbenko, Liubov Shkodenko, Maria Rubel, Aleksandr Slita, Ekaterina Nikitina, Elvira Martens, Dmitry Kolpashchikov. «DNA Nanomachine for Visual Detection of Structured RNA and Double Stranded DNA». Chemical Communications, 2022.
source (https://news.itmo.ru/en/science/life_science/news/12538/)